|
Matrices is an extension of Chemstation designed primarily to deal in a reasonable fashion with the results of time-based measurements. Matrices may be used in other circumstances, such as storing and exporting results of titrations.
Matrix is collection of multiple spectra that is handled as a single object. Even a matrix containing hundreds of spectra will appear as one line in sample table or as count of 1 in Load samples dialog. When shown in spectral overlay plots, matrix data appear in a single color, instead of a rainbow for multiple separate spectra. Selecting a matrix will sequentially highlight all entries in a matrix. Thus, selecting a 1000-spetcra matrix on a slow computer may take more than a minute.
Using matrices will involve switching between internal foldes. Review Chemstation folder structure here.
There is a limit to matrix size of approximately 2000 spectra. This limitation is likely due to antiquated memory addressing by Chemstation.
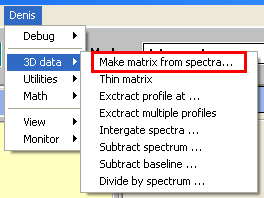 Matrix handling function appear under in menu. It is recommended, however, to limit matrix (and data in general) processing within Chemstation to a minimum and transfer bulk of to a suitable software. Matrix handling function appear under in menu. It is recommended, however, to limit matrix (and data in general) processing within Chemstation to a minimum and transfer bulk of to a suitable software.
Making matrix from spectra.
This is the primary operation that creates data matrix. To make a matrix select all the data that you want to include in a matrix and select from menu. Matrix data will appear as a new entry in MATRICES folder and original spectra will not be altered or deleted. Manually delete original set of data after confirm that matrix has been created.
When time-dependent spectral measurement is carried out, each individual spectrum contains a record of time elapsed since the experiment has started. When matrix is created from spectra, this record becomes time calibration.
|
Thin matrix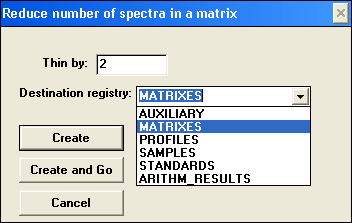
Thinning a matrix allows to reduce the number of spectra in a matrix so that only every nth spectrum is retained and spectra in between are discarded. This operation makes matrix smaller and faster to process; useful for taking an overview or snapshots of a large matrix.
Dialog specifies fraction of spectra to keep and which internal folder to put the resulting matrix to. This macro accepts one matrix at a time.
|
Extract profile at…
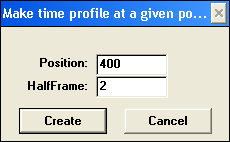 Extracting profile creates a 2-D profile of absorption change at particular wavelength over time. Results are placed in PROFILES internal folder. Extracting profile creates a 2-D profile of absorption change at particular wavelength over time. Results are placed in PROFILES internal folder.
Remember that primary calibration for profiles is time and moving any profile to any spectral window and vise versa will make viewing either one difficult.
Macro accepts one matrix at a time. Specify what wavelength should profile be centered at and what frame to average points over. For each spectrum, absorption intensities between Position-HalfFrame and Position+HalfFrame will be averaged.
|
Extract multiple profiles
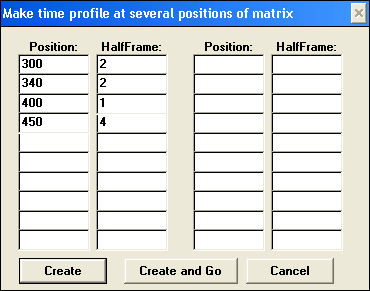 Extracting multiple profiles is equivalent to extracting profile at decribed above except that it created a matrix of profiles, i.e. several profiles at centered at different positions within one matrix. Positions and HalfFrames can be random. Resulting matrix is placed in PROFILES internal folder. Extracting multiple profiles is equivalent to extracting profile at decribed above except that it created a matrix of profiles, i.e. several profiles at centered at different positions within one matrix. Positions and HalfFrames can be random. Resulting matrix is placed in PROFILES internal folder.
|
Integrating spectra…
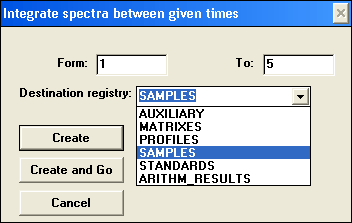 Integrating spectra over a matrix added all spectra within the matrix between From and To and divides by the number of spectra added. Limits are based on the spectrum number in the sequence, not actual the time it was taken at. Integrating spectra over a matrix added all spectra within the matrix between From and To and divides by the number of spectra added. Limits are based on the spectrum number in the sequence, not actual the time it was taken at.
|
Subtract spectrum …
Divide by spectrum…
Subtract and divide by a spectrum macros allow to subtract a 2D spectrum from a matrix or to divide a Matrix by a Spectrum. For these macros, exactly two spectra must be selected – one 3D data and one 2D data.
Resulting matrix is placed in the Matrices (or Math results?) folder.
|
Subtract baseline …
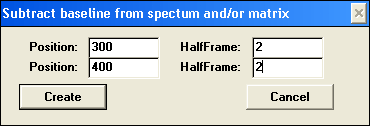 Subtract baseline macro can be used with 2D and 3D data and subtracts a linear interpolated baseline between two reference points in each spectrum within a matrix. Reference Positions and HalfFrames within which data are averaged are based on wavelength calibration, not point number. Subtract baseline macro can be used with 2D and 3D data and subtracts a linear interpolated baseline between two reference points in each spectrum within a matrix. Reference Positions and HalfFrames within which data are averaged are based on wavelength calibration, not point number.
| |


 Matrix handling function appear under
Matrix handling function appear under 
 Extracting profile creates a 2-D profile of absorption change at particular wavelength over time. Results are placed in
Extracting profile creates a 2-D profile of absorption change at particular wavelength over time. Results are placed in  Extracting multiple profiles is equivalent to extracting profile at decribed above except that it created a
Extracting multiple profiles is equivalent to extracting profile at decribed above except that it created a  Integrating spectra over a matrix added all spectra within the matrix between
Integrating spectra over a matrix added all spectra within the matrix between  Subtract baseline macro can be used with 2D and 3D data and subtracts a linear interpolated baseline between two reference points in each spectrum within a matrix. Reference
Subtract baseline macro can be used with 2D and 3D data and subtracts a linear interpolated baseline between two reference points in each spectrum within a matrix. Reference